PIPELINE MODE USAGE
3.1 - CONFIGURING AND RUNNING PIPELINES
The pipeline mode of DeNovoSeq is a pipeline configuration system that let the user to execute all the steps of a pipeline, automatically one after the other. To perform a de novo analysis using the pipeline mode, the user must access the DeNovo protocols tab of the Top menu and then select the option Pipeline Mode. Then a first pipeline interface will appear summarizing all currently available pipelines based on Command Line Interface (CLI) software offered by the Pipeline mode of DeNovoSeq.
There are three available types of pipelines;
- Genome & Transcriptome DeNovo assembly.
- Augustus-based Gene Prediction
- NCBI-Blast-based Annotation
To perform an analysis using the pipeline mode, please go to [ Transcripts Protocols → Pipeline Mode ] and follow Fig. 37
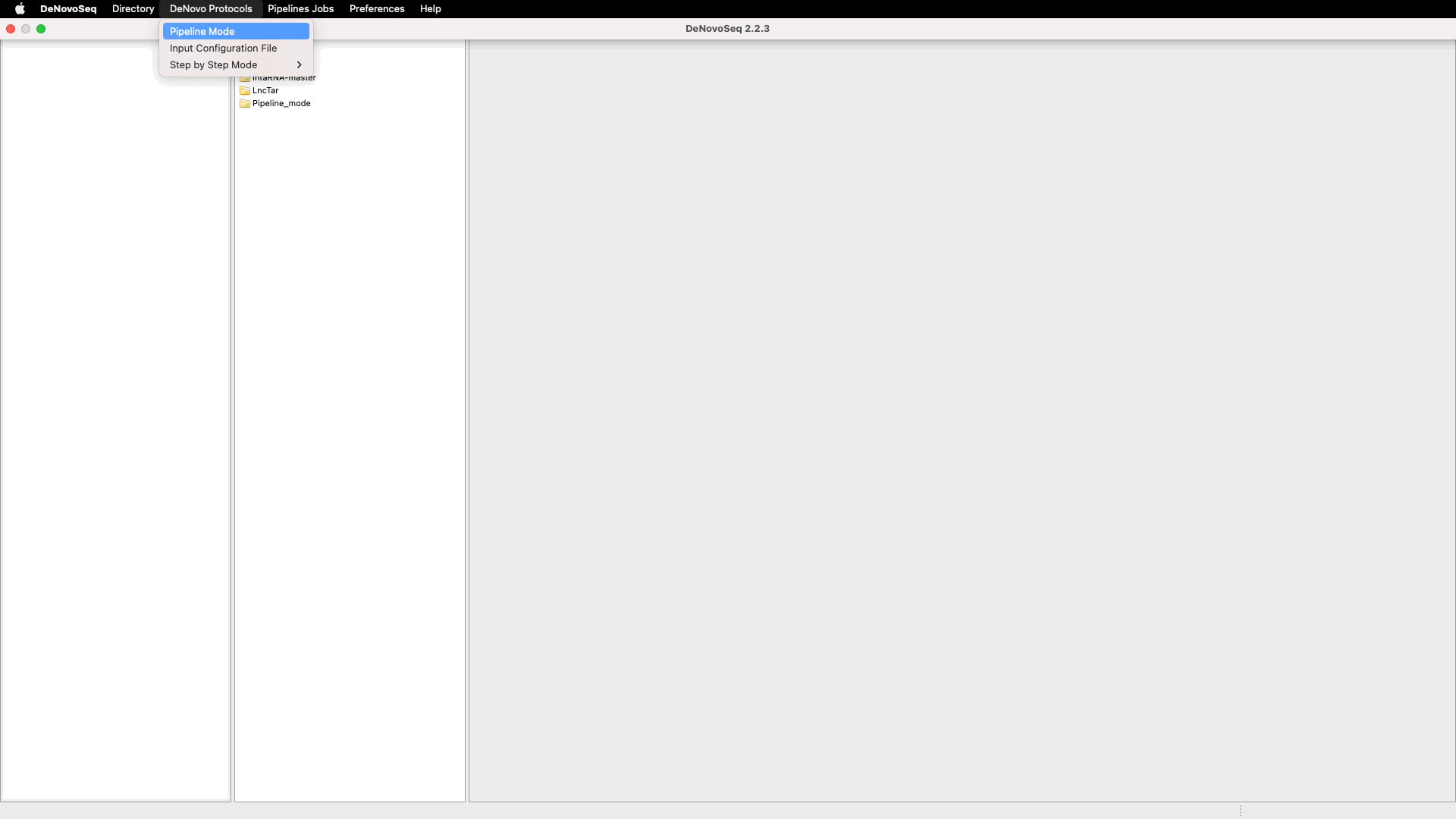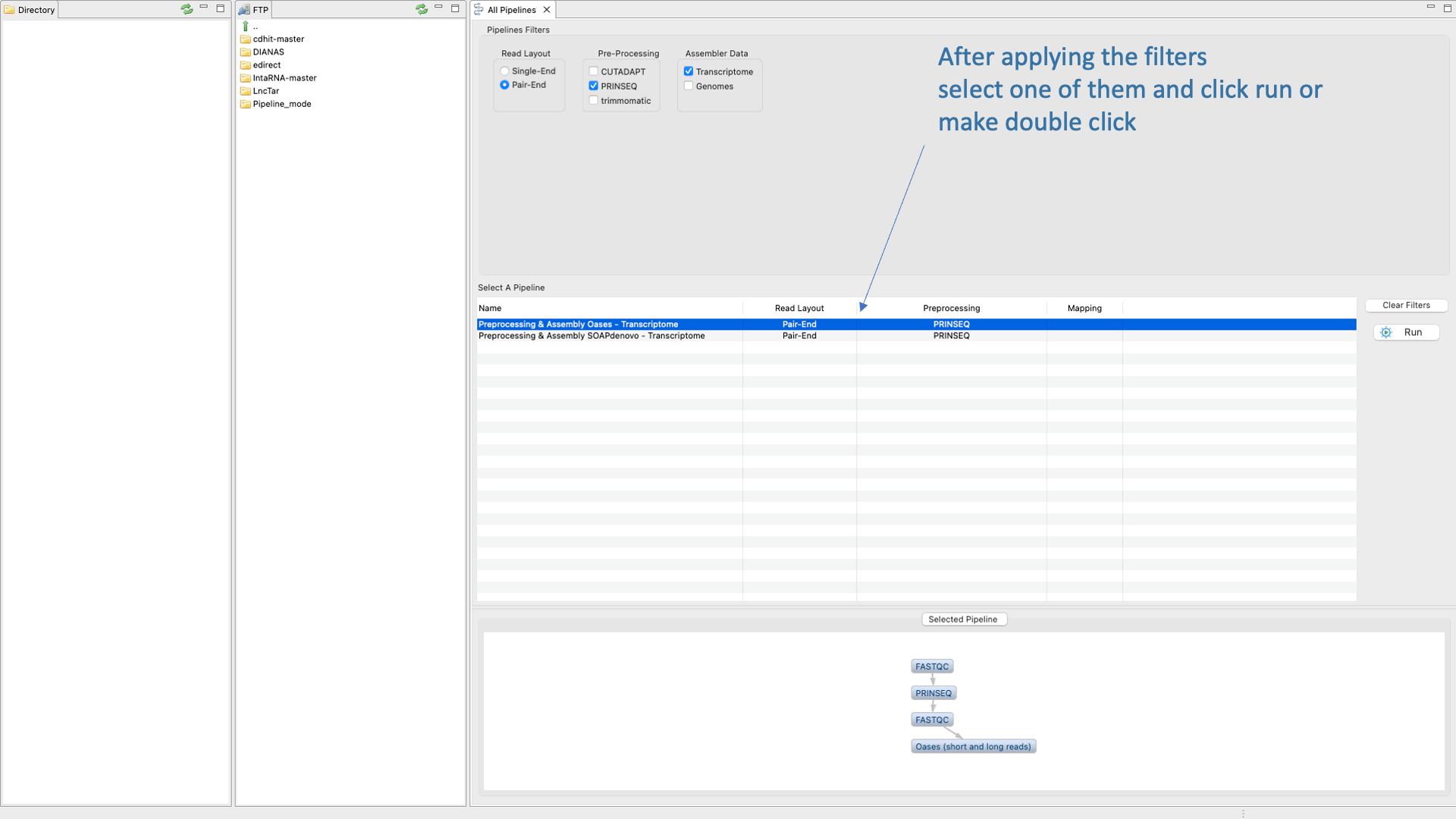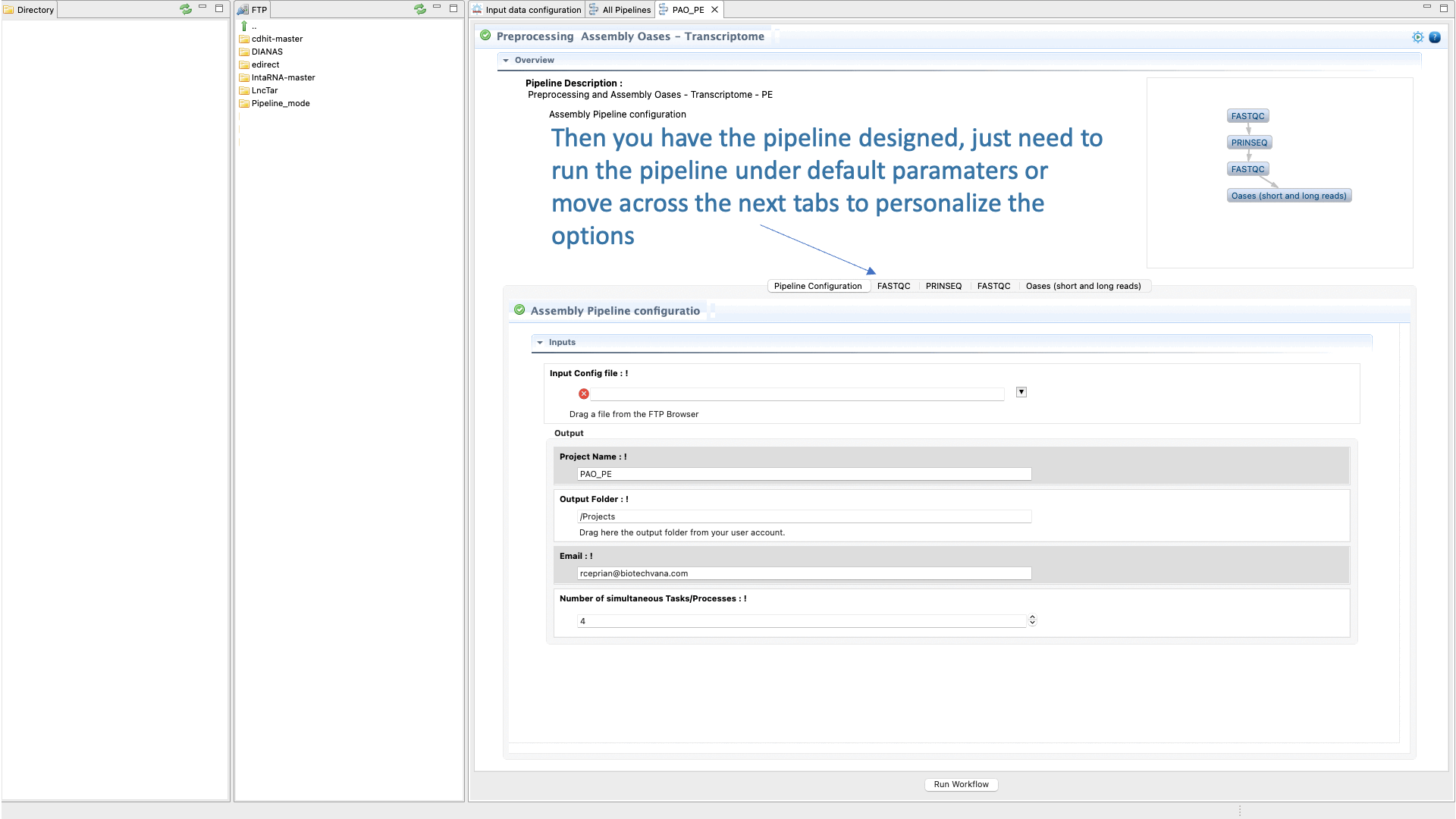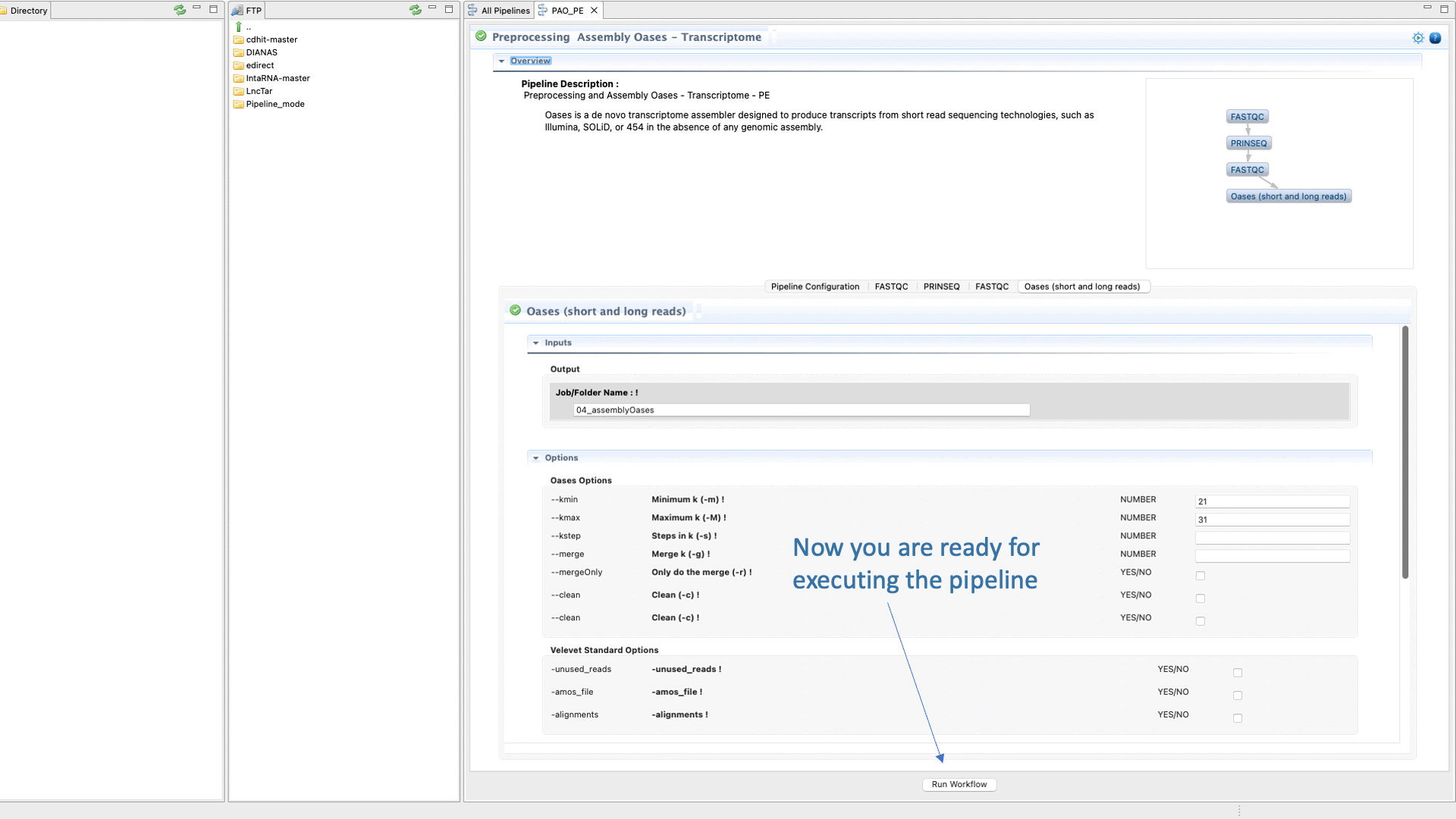
Figure 37: Example of configuration and execution for a DeNovoSeq analysis using the pipeline mode.
Using the pipeline mode you only need to select one pipeline from the list and then configure the experiment going to a set of nested interfaces to;
- Upload any input data or material needed by the pipeline (i.e. fastq files, input data configuration.
- Declare the outputs
- Configure the parameters and options of each CLI tool used in each step.
- Finally run the pipeline.



