
La digitalización in silico de escenarios de mundo real mediante gemelos digitales y simulaciones avanzadas permite comprender la dinámica interna de cualquier sistema bajo estudio y predecir su comportamiento a lo largo del tiempo. Gracias a estas herramientas es posible evaluar cómo evoluciona un sistema bajo condiciones naturales o cómo responde ante cualquier tipo de intervención inducida, superando las limitaciones de los modelos predictivos convencionales. Esto facilita la optimización de experimentos, el ahorro de recursos y la generación de conocimiento predictivo en múltiples ámbitos de las ciencias de la vida y más allá.
Para construir los simuladores que recrean el gemelo digital de un caso de estudio empleamos diversas tecnologías, con mención destacada a la tecnología de Membrane Computing, un paradigma de computación natural inspirado en la organización y funcionamiento de los sistemas biológicos. Este enfoque permite modelar procesos complejos mediante dinámicas con niveles anidados de complejidad (molecular, celular, tisular, individual, poblacional, etc.), en los que los fenómenos que ocurren en un nivel pueden repercutir en otros de mayor jerarquía.
Este tipo de aproximaciones hace posible explorar escenarios de naturaleza muy diversa para evaluar respuestas a intervenciones, optimizar sistemas complejos bajo entornos controlados y seguros y generar predicciones detalladas imposibles de alcanzar mediante métodos tradicionales.
Se recopilan datos de sensores, bioensayos, experimentos ómicos o mediciones de laboratorio, creando una base sólida para la representación digital del sistema real.
Los datos se integran en modelos computacionales (en el caso de Membrane Computing son conocidos como sistemas P) o algoritmos que reproducen los procesos a simular.
Mediante inteligencia artificial y aprendizaje automático, el gemelo digital identifica patrones, predice resultados y ayuda a tomar decisiones más rápidas y precisas en investigación, desarrollo o producción.
Anticipa resultados experimentales, variaciones genéticas o cambios en procesos biológicos complejos mediante simulaciones de alta precisión.
Reduce tiempos y costes de ensayo al probar condiciones, parámetros o tratamientos de forma virtual antes de implementarlos en laboratorio o producción.
Permite iterar hipótesis rápidamente, generar conocimiento predictivo y acortar los ciclos de desarrollo científico o biotecnológico.
Conecta modelos predictivos y analítica avanzada para apoyar decisiones estratégicas en investigación, salud, agricultura o bioindustria.
Combina machine learning, modelado molecular y simulación de datos ómicos para construir ecosistemas biológicos digitales conectados.
Entre los sistemas que podemos modelar se incluyen: dinámicas de resistencia microbiana, enfermedades genéticas, respuestas metabólicas e inmunes frente a infecciones, fármacos o vacunas, ecosistemas naturales y sociales, procesos industriales, fenómenos meteorológicos extremos (DANAs, terremotos, incendios), pandemias y epidemias, escenarios de defensa estratégica y modelos de riesgo, entre muchos otros.
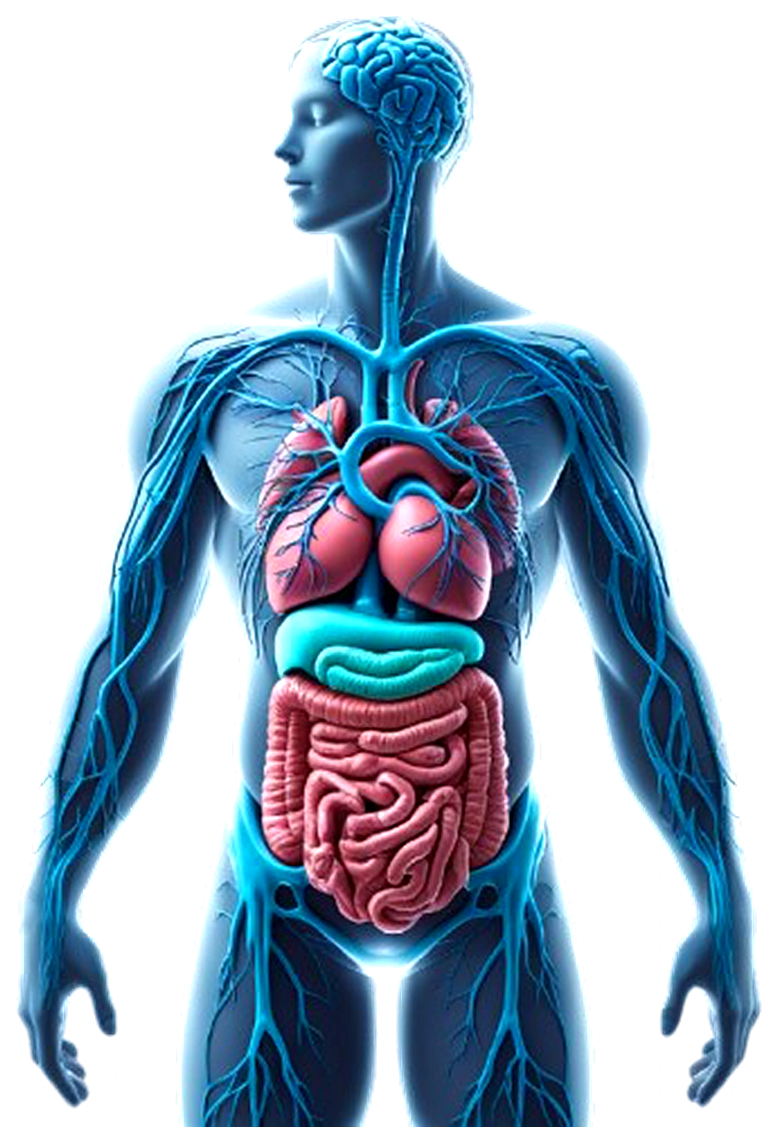
El gemelo digital permite recrear de manera virtual y altamente precisa el cuerpo humano, generando un modelo que imita tanto su estructura física como sus funciones biológicas. A través de esta simulación es posible analizar, predecir y visualizar cómo respondería el organismo ante la aplicación de una vacuna antes de llevarla a cabo en la realidad.
- 1
- 2
- 3


El gemelo digital permite recrear digitalmente el cuerpo humano para analizar y predecir cómo responde a una vacuna antes de su aplicación real.
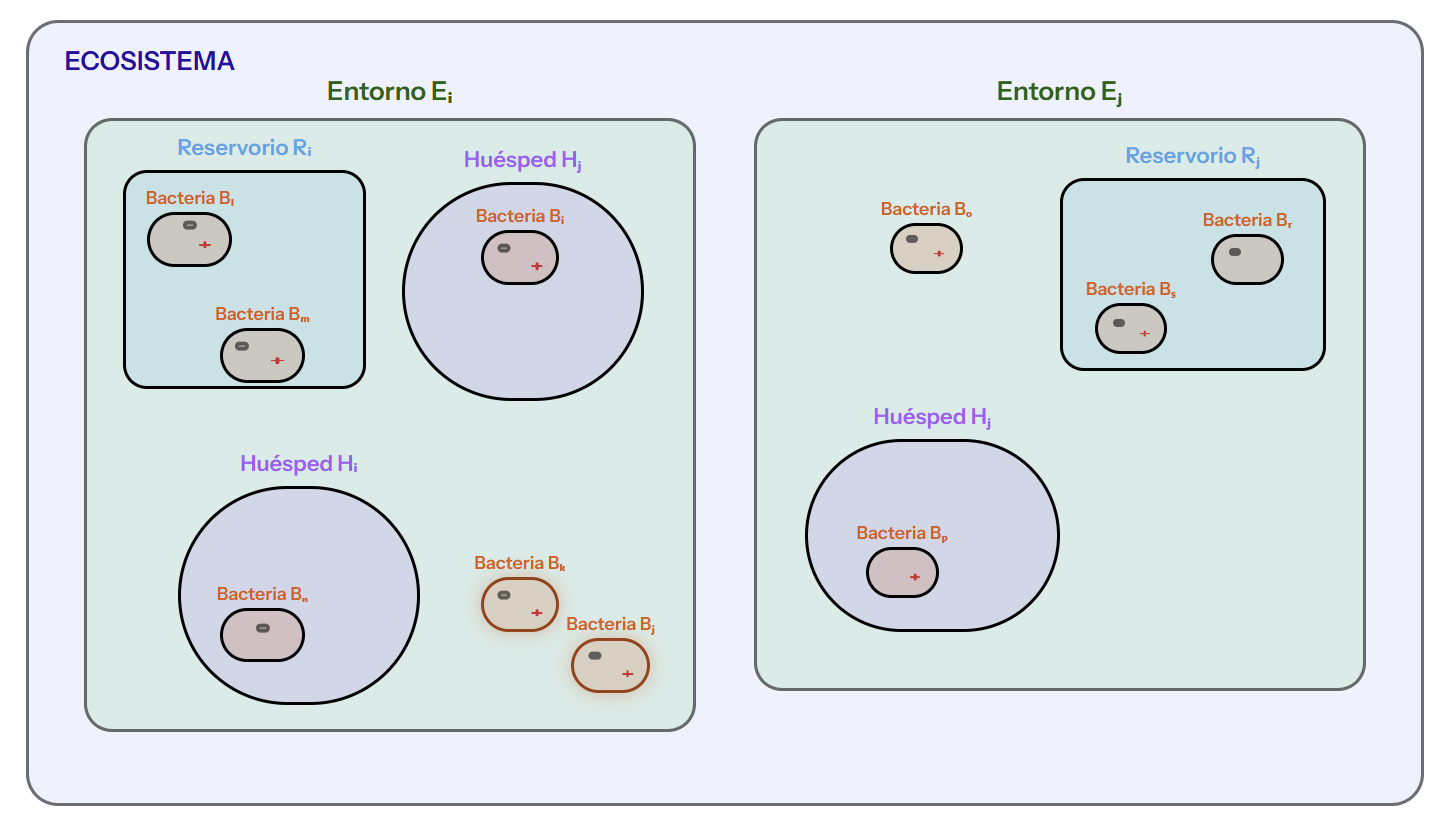
El gemelo digital simula la dinámica de genes de resistencia, plásmidos y bacterias a través de distintos niveles del ecosistema (huéspedes, reservorios y entornos), facilitando la evaluación de riesgo y el diseño de estrategias de control sin necesidad de ensayos directos en el mundo real.
Si estás interesado en obtener más detalles sobre Digital Twins, por favor contáctanos en biotechvana@biotechvana.com para valorar una solución hecha a medida para tu proyecto.
Contáctanos


