Features
CheckAlign generates graphical representations of DNA or protein multiple alignments sequences, creating outputs in two formats: PDF and PostScript.
The update of CheckAlign 2.0 adapts the logo maker to the most common operating systems updates and additionally focused on an implementation of the Shannon-Weaver algorithm to characterize species diversity in a community.

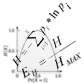
Logo obtained using Shannon’s algorithm with standard deviation bars
Estimating diversity index measures for one or more biological or molecular samples
Users may build the logo from a gapped alignment considering the gap as another nucleotide or amino acid species; moreover they can use the correction factor (recommended for small alignments) and depict standard deviation bars. Note that the “frequency algorithm” option creates a logo but the representation results in an overestimation of the true consensus.
 Logo obtained using Shannon’s algorithm with standard deviation bars
Logo obtained using Shannon’s algorithm with standard deviation bars
 Estimating diversity index measures for one or more biological or molecular samples
Estimating diversity index measures for one or more biological or molecular samples



